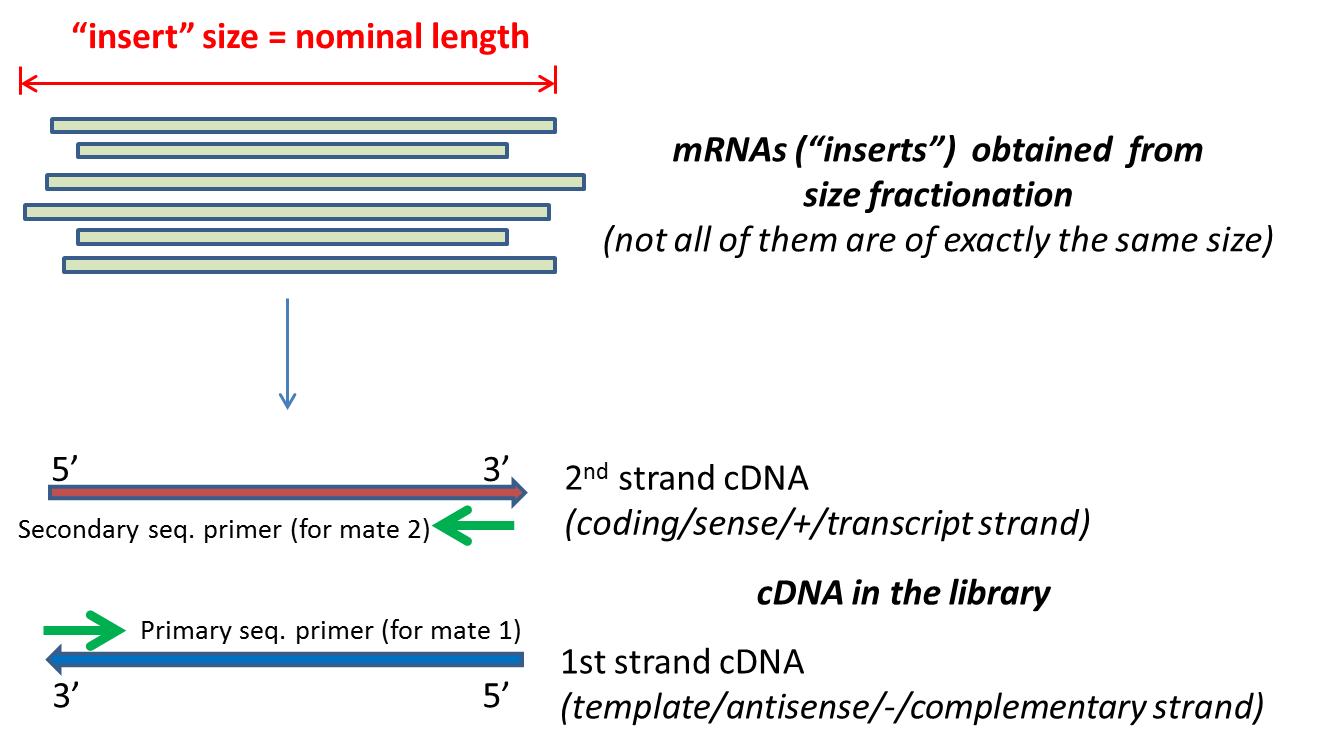
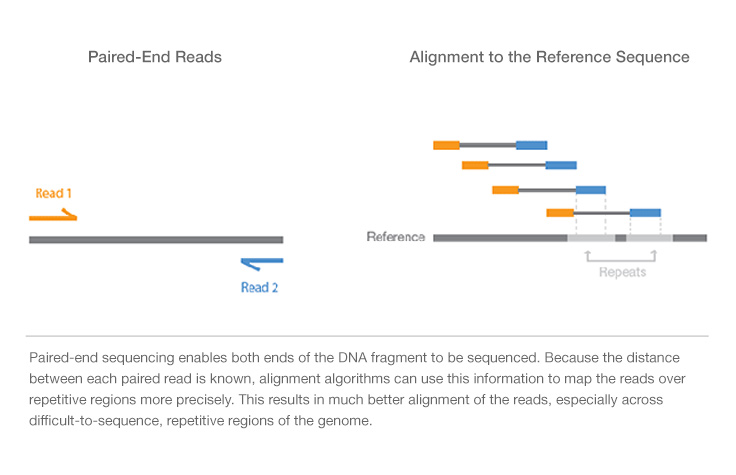
paired end sequencing insert size
For instance for a PE150 sequencing run the insert size should be above 300bp. The inner mate distance.
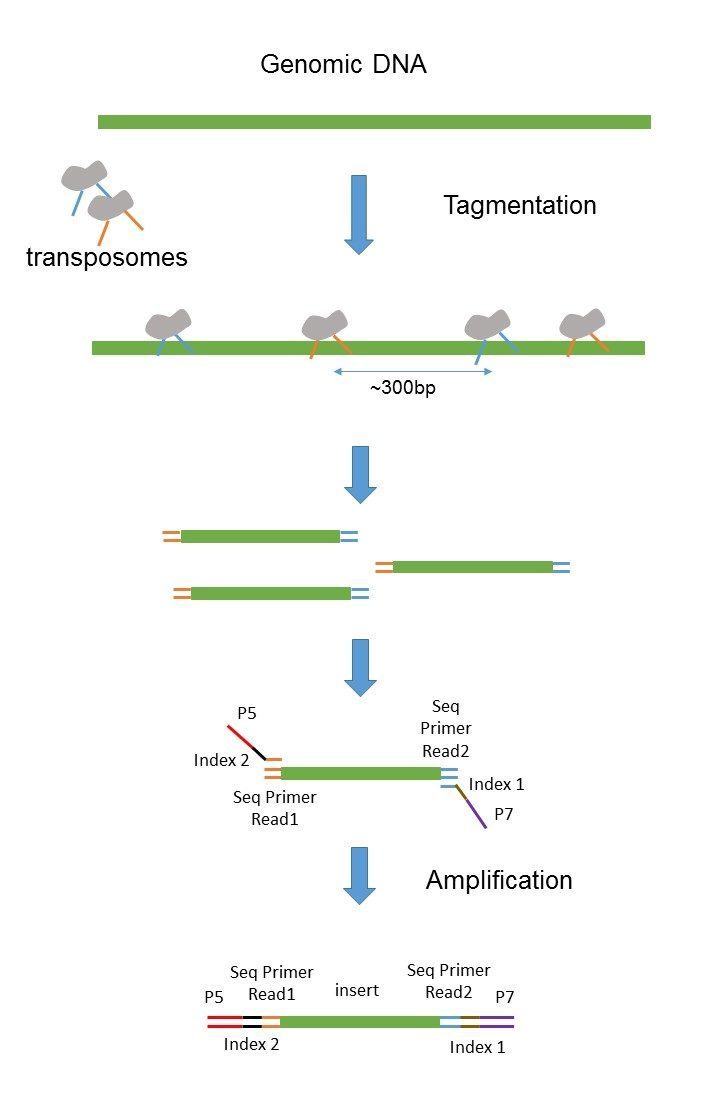
Paired End Sequencing France Genomique
This can result in a proportion of fragments with an insert size of less than the length of a.

. Read files from paired-end sequencing need to be paired in Geneious before the pairing information can be used in assembly. Read length of paired-end reads. Mean 29622 STD2251 Read span.
The sequencing starts at Read 1 Adapter mate 1 and ends with the sequencing from Read 2 Adapter mate 2. Simple workflow allows generation of unique ranges of insert sizes. The CollectInsertSizeMetrics tool outputs the percentages of read pairs in each of the three.
For the first test I took some sequence from the human genome hg19 and created two 100 bp reads from this region. The menu function Tools Genome Utilities llumina Insert Sizes Picard can be used to analyze the distribution of insert sizes of aligned reads. The reads have a length of typically 50 - 300 bp.
Standard paired-end libraries 200500 bp can be used to detect large and small insertions deletions indels inversions and other rearrangements. This can be done using the Set Paired Reads. The variation of insert sizes is often large and the average size difficult to control.
The insert size on classic paired-end is smaller about 500bp while the insert size of mate-pair is much longer several Kb which allows to join the. Despite the benefits of gaining SS-PE data with paired ends of varying distance the standard Illumina protocol allows only non-strand-specific paired-end sequencing with a single insert. Including flanking intronic regions of.
The insert is normally the stretch of sequence between the paired-end adapters so in your case the insert size would be 250 bp 2x75 bp reads 100 bp unsequenced middle. These reads can then be sequenced using the same SP1-SP2 adapter protocols used in PE sequencing. Quality of the second read in paired-end sequencing.
I got this output. For classical paired-end. Fast and Accurate Next-Generation Sequencing Results Enabled by Ion Torrent Technology.
If 150 bp is each single end length then fragment size can not be 250 inner distance can not be negative if 275 is 150 then insertion size you should provide is 100 bps. Ad Gene Expression Profiling Chromosome Counting Epigenetic Changes Molecular Analysis. 2Align paired-end RNA-Seq data to the reference transcripts using minimum insert-length as zero.
Ad Gene Expression Profiling Chromosome Counting Epigenetic Changes Molecular Analysis. Insert size -- The insert size refers to the distance between the pairs. Mean 48454 STD10836 Does it mean my insert size is 484-296184.
When invoked the SAMBAM file that should be. Requires the same amount. Now lets get started.
The insert size on classic paired-end is smaller about 500bp while the insert size of mate-pair is much longer several Kb which allows to join the contiguous between them. Paired-end sequencing also provides. To read more about the different parts of a prepared DNA fragment please take a look on the figure in the article about observed sequence lengths in Illumina.
Fast and Accurate Next-Generation Sequencing Results Enabled by Ion Torrent Technology. Paired end sequencing refers to the fact that the fragments sequenced were sequenced from both ends and not just the one as was true for first generation sequencing. Nine percent of read pairs were found to have an insert size of less than 250 bp and so likely to contain adapter after mapping to the reference genome of which 98 were also found to have.
1Create reference transcriptome index for BowTie or BWA. However the average size of human coding exons is only 160bp. For a brief primer on paired-end sequencing and mate-pair reads see the GATK Dictionary.
We surveyed publicly available paired-end sequencing data downloaded from Illumina BaseSpace and SRA that. Rob Edwards from San Diego State University describes how Illumina paired-end sequencing works.

What Are Paired End Reads The Sequencing Center
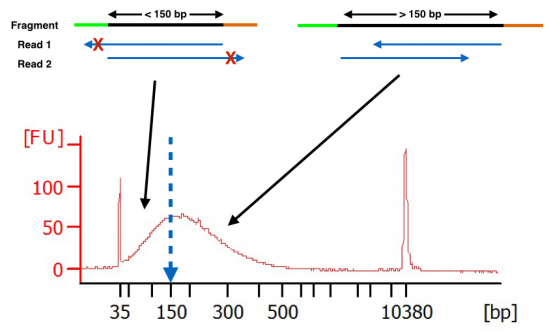
What Happens When Hardware Advances Faster Than Molecular Protocols Cofactor Genomics

Interpreting Color By Insert Size Integrative Genomics Viewer

What Is Mate Pair Sequencing For
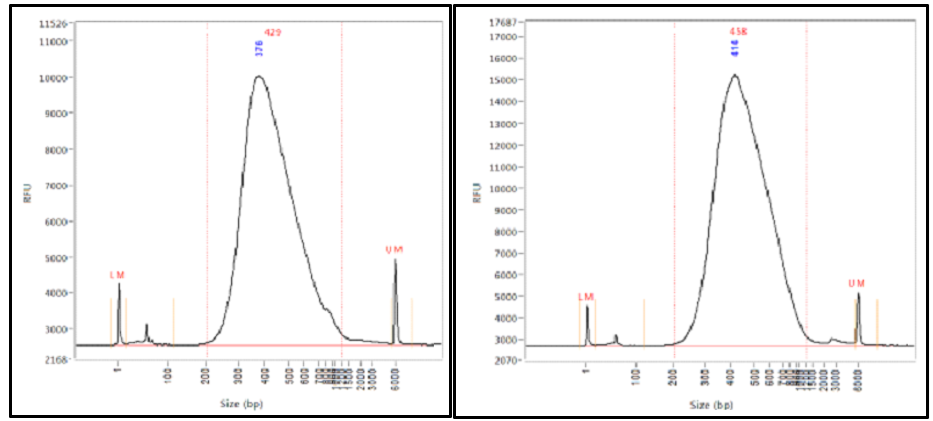
How Short Inserts Affect Sequencing Performance

What Are Paired End Reads The Sequencing Center

Diagram To Show The Construction Of A Fragment With An Insert Size Is Download Scientific Diagram
What Read Lengths Are Produced By Modern Illumina Sequencers

Diagram To Show The Construction Of A Fragment With An Insert Size Is Download Scientific Diagram
The Insert Size In Paired End Data Seqanswers

Pdf Assessment Of Insert Sizes And Adapter Content In Fastq Data From Nexteraxt Libraries
What Is Paired End 150 Omega Bioservices

What Is Mate Pair Sequencing For
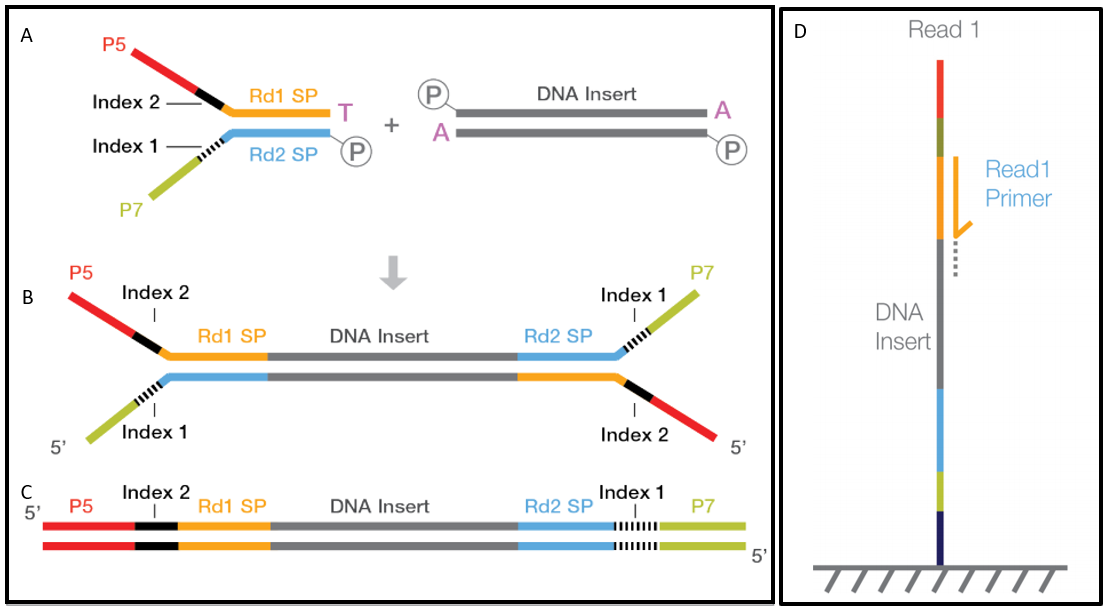
How Short Inserts Affect Sequencing Performance

Library Insert Size Libraries Were Constructed Using The Standard A Download Scientific Diagram

Definition Of Essential Terms A Definition Of Fragment Read And Download Scientific Diagram

Interpreting Color By Insert Size Integrative Genomics Viewer